How to Read an Xml File in Python
Python classical way of handling big XML files
Logically reading the XML file in python
Python — It doesn't matter nosotros're beginners or gurus, Python gives united states a vast variety of libraries, methods, and functions as per our demand. Isn't it?
When it comes to reading a large file, nosotros all recall how much memory and time our program going to consume to do the piece of work and provide expected results. Either Data Scientific discipline or Machine Learning, cleaning a large set of data with a minimal amount of time is the cardinal to the success of building complicated algorithms.
There are a lot of modules we can detect in Python to read the XML, JSON, YAML and Flat text files. Especially if we demand to read a huge size of an XML file, python modules utilize quite an corporeality of time to construct the root, child element, and subsequent elements. Every bit the file grows our code started performing slow and takes fourth dimension to consummate.
The issue or business concern hither is not with the python module, but with the volume of data . Below can be an culling approach to handle the huge volume of information with less execution fourth dimension.
What are the modules we can attempt parsing XML files?
- xml.etree — Simplest way of processing the xml files and it'south lightweight.

- xml.dom.minidom — Minimal implementation of the Document Object Model (dom) interface.

- xml.dom.pulldom — Outcome-driven processing model, explicitly pulling events from the stream until all effect processing completes or an fault occurs.

- xml.sax — Unproblematic API for xml processing.

How do nosotros measure out the operation of XML file processing?
In real world scenarios, well-nigh of the time we read the larger XML file, merely we might need but a few blocks of elements to compute the results.
Let's take an instance of below XML file, consider there are 500k ProteinEntry elements in the XML file and file size is around 650 MB or sometimes we try to process more than that. All we need is the <header> block from this huge XML file.
Sample XML Block:
<ProteinEntry id="123">
<header>
<uid>CCCZ</uid>
<accession>A00002</accession>
<created_date>17-Mar-1987</created_date>
<seq-rev_date>17-Mar-1987</seq-rev_date>
<txt-rev_date>03-Mar-2000</txt-rev_date>
</header>
<protein>
<proper noun>cytochrome c</name>
</poly peptide>
<organism>
<source>chimpanzee</source>
<mutual>chimpanzee</mutual>
<formal>Pan troglodytes</formal>
</organism>
<reference>
<refinfo refid="A94601">
<authors>
<writer>Needleman, Southward.B.</author>
</authors>
<citation type="submission">submitted to the Atlas</citation>
<calendar month>October</month><twelvemonth>1968</year>
</refinfo>
<accinfo label="NEE">
<accession>A00002</accession>
<mol-type>protein</mol-type>
<seq-spec>1-104</seq-spec>
</accinfo>
</reference>
</ProteinEntry> Permit's create a method and measure it. Remember all we need is header count and header content from the XML file. We are going to use xml.etree.cElementTree to read the XML file(for testing purpose).
1. Get the file size
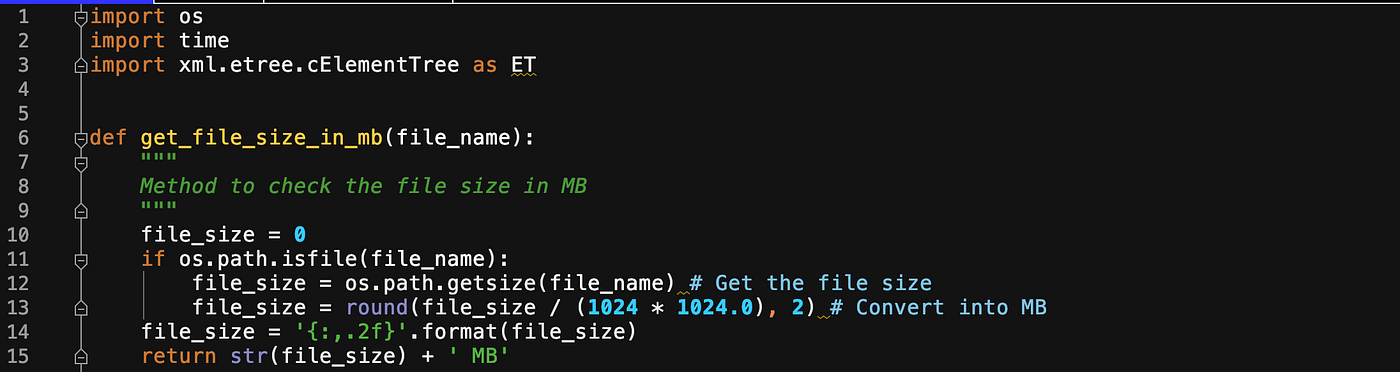
2. Process the file through xml.etree

3. Identify the processing time
Results with xml.etree.ElementTree Reader:
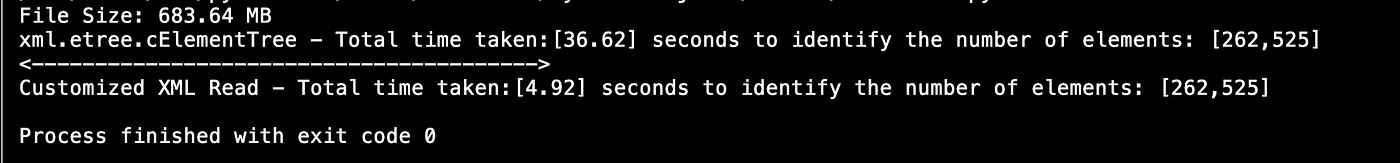
It took 34.11 seconds to read 683 MB of XML file, and there are 262K headers we captured.

4. Writing customized XML processor
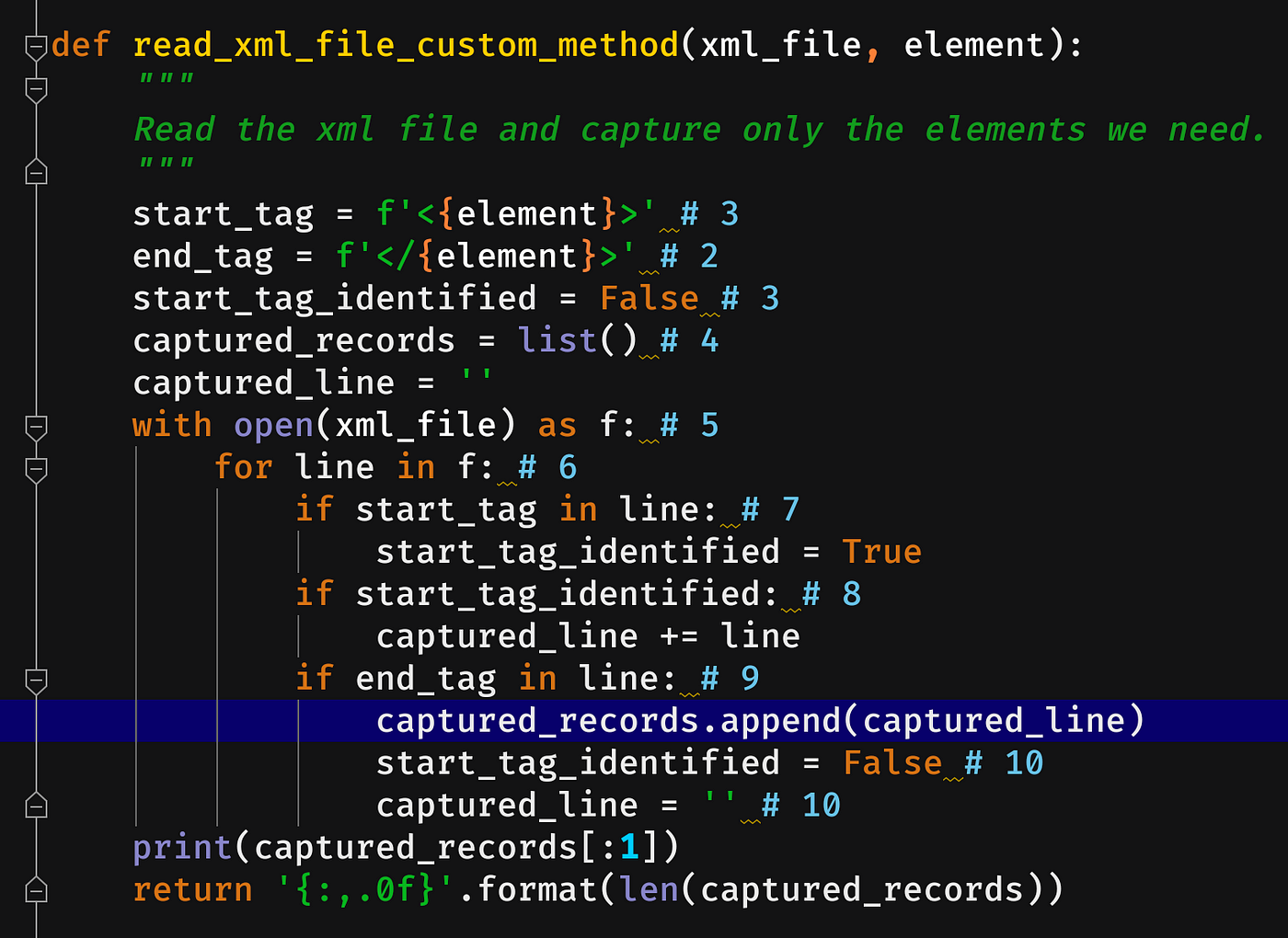
Steps:
- Element start-tag (<header>)
- Element end-tag (</header>)
- Indicator start_tag_identified to track the starting element. In one case identified mark it equally True.
- Captured elements volition exist stored in a list — captured_records
- Read the XML file
- Process the file line-by-line
- Identify the start tag and modify the indicator to True — start_tag_identified
- Once the start-tag is identified, capture the lines between them.
- Once we reached the end-tag, capture all lines betwixt the start and end tag then shop it in a list.
- Reset the values to move to the next set of the element (<header> … </header>)
Time to test our customized XML Reader Code:

Results with customized XML Reader Code:
All it took only 4.92 seconds to read the 683 MB of file and there are same 262K headers nosotros catpured . We are able to save virtually thirty–33 seconds of processing time. I would say execution time plays a key role when information technology comes to process multiple files concurrently (similar multi-threading).
Code To Refer:
import os
import fourth dimension
import xml.etree.cElementTree as ET
def get_file_size_in_mb(file_name):
"""
Method to cheque the file size in MB
"""
file_size = 0
if bone.path.isfile(file_name):
file_size = bone.path.getsize(file_name) # Get the file size
file_size = round(file_size / (1024 * 1024.0), 2) # Convert into MB
file_size = '{:,.2f}'.format(file_size)
return str(file_size) + ' MB'
def read_xml_file(xml_file, element):
"""
Parse the xml file to xml.etree.cElementTree
"""
tree = ET.parse(xml_file)
root = tree.getroot()
number_of_element = len(root.findall(chemical element))
render '{:,.0f}'.format(number_of_element)
def read_xml_file_line_basis(xml_file, chemical element):
"""
Read the xml file and capture only the elements we need.
"""
start_tag = f'<{chemical element}>' # 3
end_tag = f'</{chemical element}>' # 2
start_tag_identified = Fake # three
captured_records = list() # four
captured_line = ''
with open up(xml_file) every bit f: # v
for line in f: # 6
if start_tag in line: # 7
start_tag_identified = True
if start_tag_identified: # 8
captured_line += line
if end_tag in line: # 9
captured_records.suspend(captured_line)
start_tag_identified = Fake # 10
captured_line = '' # x
render '{:,.0f}'.format(len(captured_records))
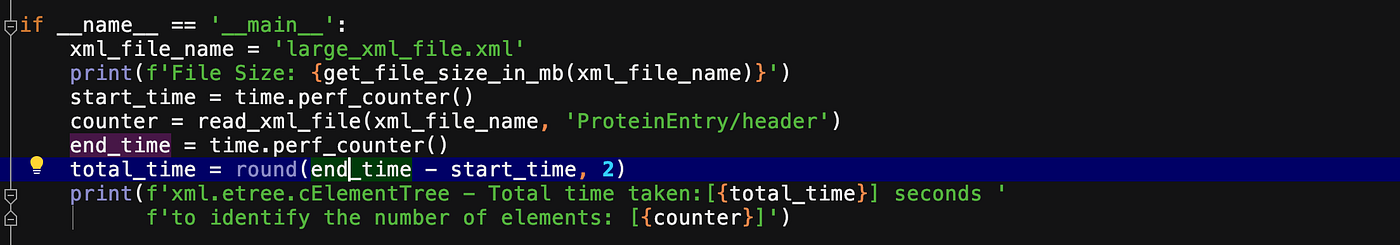
if __name__ == '__main__':
xml_file_name = 'large_xml_file.xml'
print(f'File Size: {get_file_size_in_mb(xml_file_name)}')
start_time = time.perf_counter()
counter = read_xml_file(xml_file_name, 'ProteinEntry/header')
end_time = time.perf_counter()
total_time = round(end_time - start_time, 2)
print(f'xml.etree.cElementTree - Total time taken:[{total_time}] seconds to identify the number of elements: [{counter}]') print("<---------------------------------------->") start_time = time.perf_counter()
counter = read_xml_file_line_basis(xml_file_name, 'header')
end_time = time.perf_counter()
total_time = round(end_time - start_time, 2)
print(f'Customized XML Read - Total time taken:[{total_time}] seconds to identify the number of elements: [{counter}]')
Determination:
Every bit the saying goes…
— Decision-making complexity is the essence of computer programming.
To answer the questions we accept with our code is not always going to be existing divers ways, but we have to build our own logical ways to overcome that.
Source: https://medium.com/@ggopi19/python-classical-way-of-handling-large-xml-files-e7f86e95e6f9
0 Response to "How to Read an Xml File in Python"
Post a Comment